
January 2022
Bovine Neonatal Diarrhea Panel Results
By Dr. Gregg Hanzlicek
This is a descriptive summary of the pathogens identified by PCR testing on bovine diarrhea submissions to KSVDL.
In 2021, 439 samples were submitted. Of those submissions, 61.5% contained at least one known associated pathogen. There was no difference in the percentage of positive samples by sample type, including swab, feces or intestinal tissue.
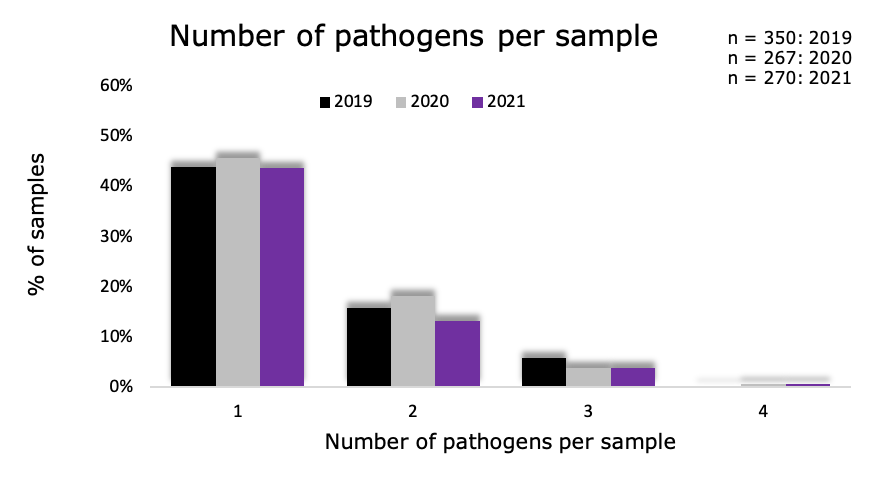
Graph 1. Of the positive samples, 43.7% contained one pathogen. Few samples contained three or more pathogens.
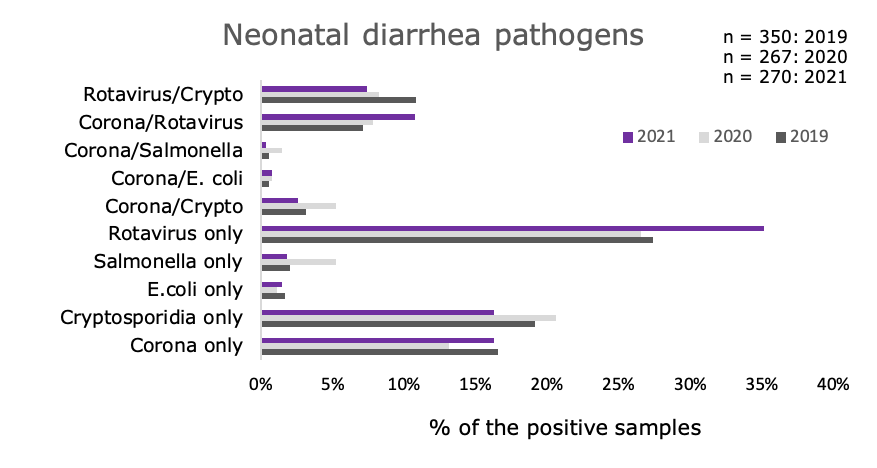
Graph 2. More samples contained Rotavirus alone (35.2%) than any other pathogen. Cryptosporidia and Coronavirus were tied as the second most commonly found single pathogens (16.3%). Rotavirus identified with either Cryptosporidia or Coronavirus were the most common combination pathogens, 7.4% and 10.7%, respectively.
Salmonella, was found in only a small percentage of samples, 1.9% alone and 0.4% with Coronavirus, its presence should not be discounted as it represents one of the two zoonotic pathogens, the other being Cryptosporidia.
KSVDL now offers a sequencing service that provides the ability to discover the level of genetic similarity between a pathogen found in the diarrhea sample and vaccine that is being used on that particular operation. A great genetic dissimilarity does indicate the potential for less than desirable cross protection provided by the vaccine strain for the strain present on the producer’s operation.
Graph 1:
 Graph 2:
Graph 2:
NEXT: Now Accepting Bovine Trichomonas Samples Submitted in Saline
Return to Index